
- #Cytoscape node attributes install
- #Cytoscape node attributes update
- #Cytoscape node attributes download
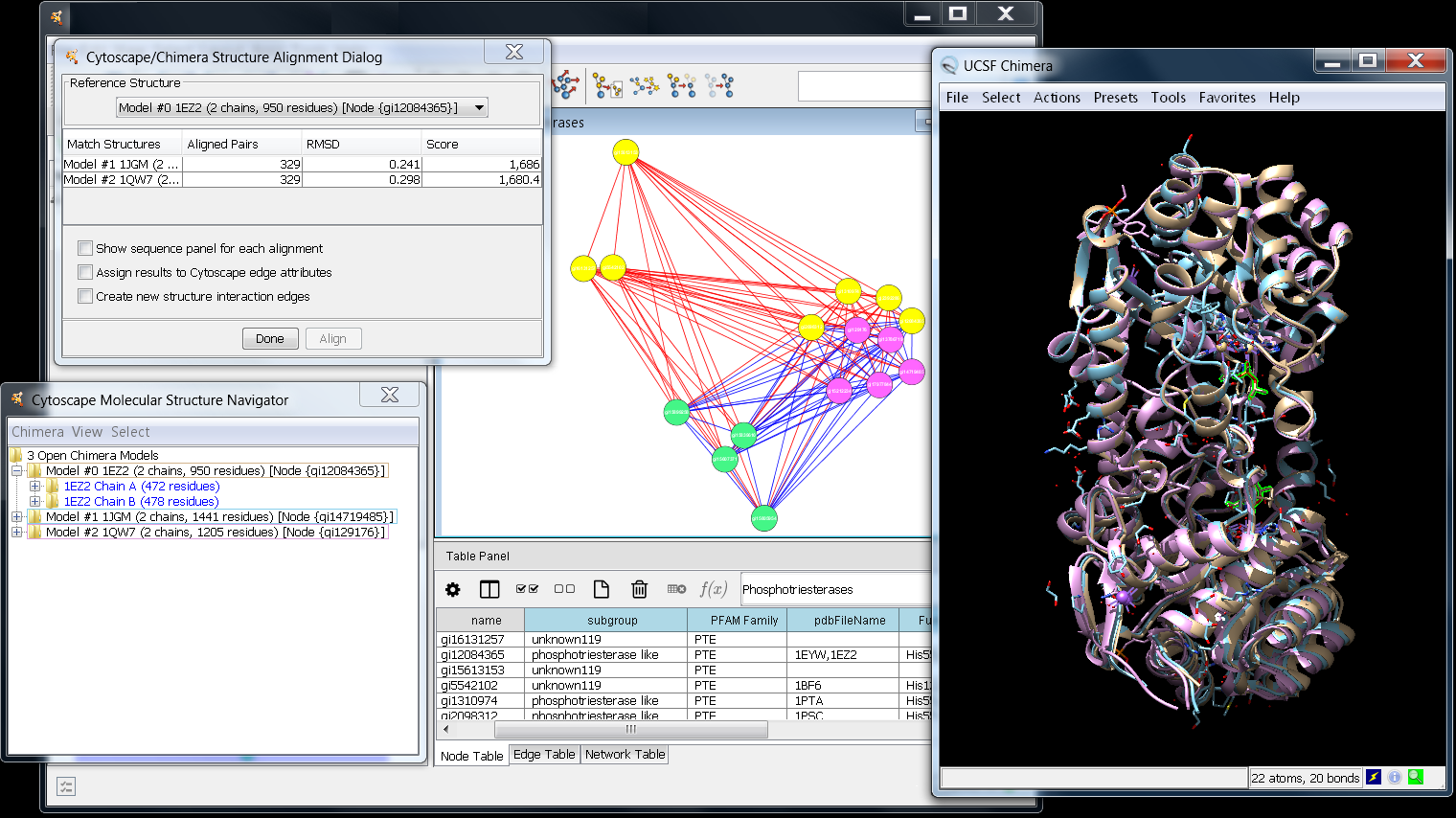
The MCODE sub-menu provides two additional options: The main MCODE interface will appear as a tab in the left-hand panel of Cytoscape (Note: If MCODE does not appear in the Plugins Menu, then revisit the installation section above.).
#Cytoscape node attributes update
Otherwise, this may happen when Cytoscape releases an update and MCODE's compatibility information becomes outdated. If you do not see MCODE in this section, it may be because the plugin is already installed and no new versions have come out since the last installation.
#Cytoscape node attributes install
Go to Available for Install > Analysis and select MCODE. Automated Installation via the Plugin Manager (available with Cytoscape 2.5 or later) Once you have downloaded and installed Cytoscape and verified that it works, MCODE can be installed in two ways.
#Cytoscape node attributes download
You can download a copy of Cytoscape from.
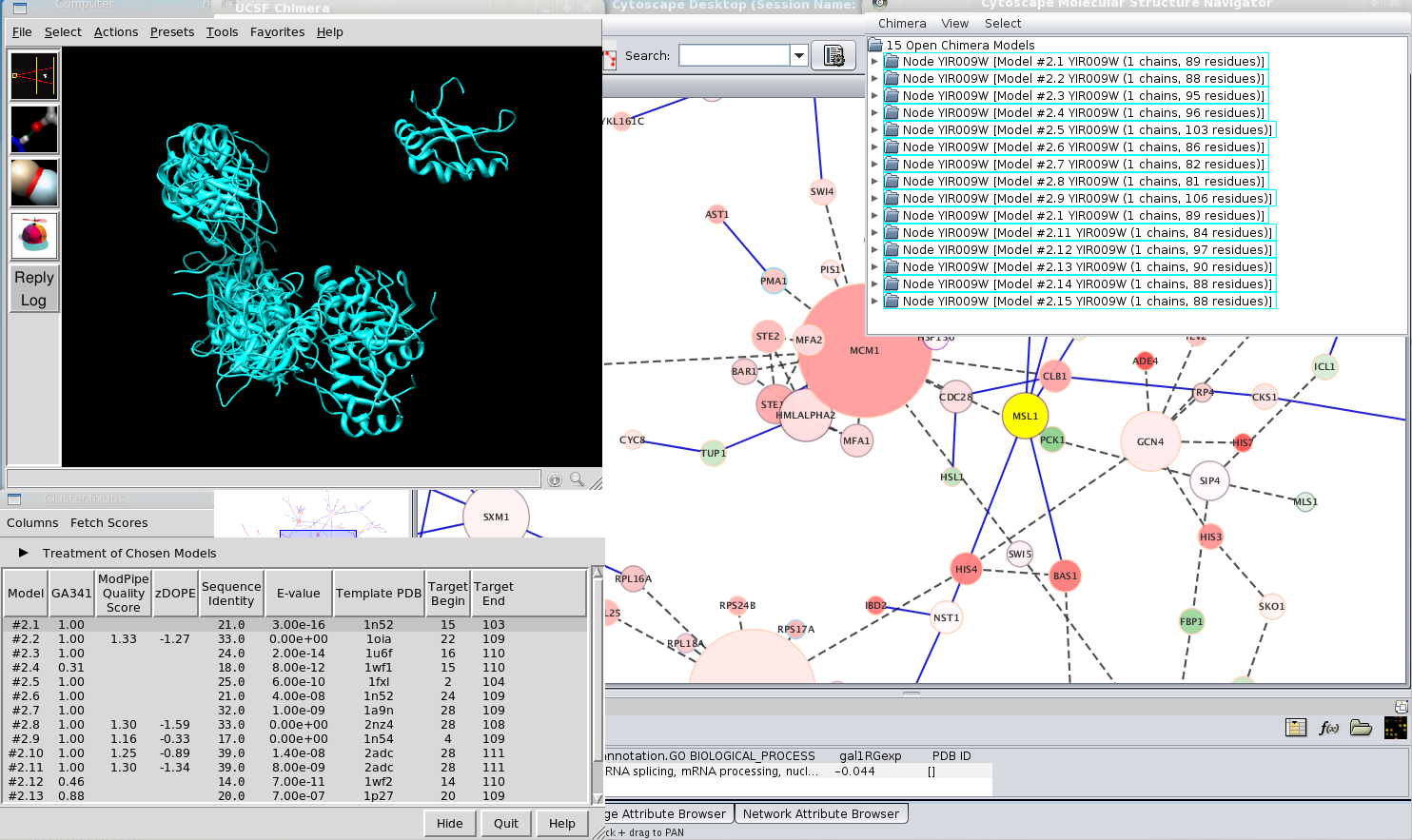
The latest MCODE, version 1.3, requires Cytoscape 2.5.1 or later. The compatible MCODE and Cytoscape versions are outlined in the downloads section on the MCODE website. If you only interested in interactions among given genes, you can do this by following “Loading a network” and you need to check the box of “Search Paths Between Genes”.To use the MCODE plugin, you must first obtain and install Cytoscape.It has three levels from 1 to 3 that mean users can choose different number of steps of expansion to find neighbors. There is interaction level drop-down list on the dialog box.Right click a node/edge->annotation->log in->change annotation set name->edit attributes from MiMI, add comments and URLs->save->log out.Select nodes and edges->right click node->SAGA->apply SAGA to chosen network->specify database and matching percentage->query.Click “sort” to sort the result with MEADįinding pathway matches for chosen network Right click an edge->get literature info from BioNLP.If you need to load a network, click here. Assuming that you already loaded a network.Getting literature information for selected edge (interaction) and sort the result Double clicking node/edge will direct you to MiMI webpage for that node/edge.Browse node/edge attributes with Cytoscape Node Attribute Browser and Edge Attribute Browser.Ī) Click Node Attribute Browser on Cytoscape panel (bottom right)->click icon ->check attributes name->choose nodes to see attributes.ī) Click Edge Attribute Browser to get edge information.Ī) Starting up Cytoscape->plugins->MiMI->MiMI Plugin->From File tab-> Drop down box ->specify species, molecule type etc Prepare a list of genes, see, for example mygene.txt.If you need to install Cytoscape, click here. Assuming that you already installed Cytoscape and MiMI Plugin.Finding pathway matches for chosen networkĬlick here to get instructions.Getting literature information for selected edge and sort the result.


 0 kommentar(er)
0 kommentar(er)
